Predicting biophysical characteristics of proteins from their amino acid sequence
Category
Structural BioinformaticsAbout This Project
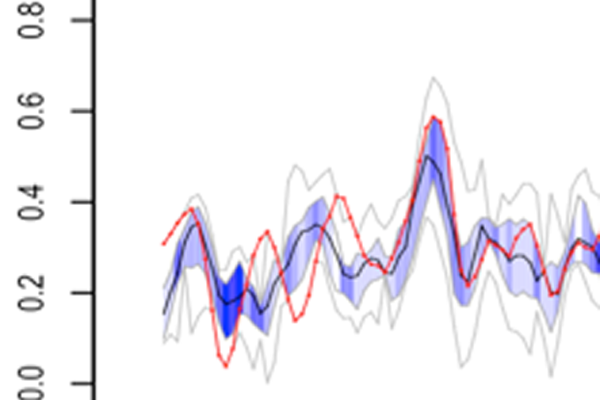
Most methods that predict per-residue characteristics from sequence are trained on information from in silico protein structures, which do not necessarily capture how the protein behaves in solution (e.g. helix fraying, loop movements). We are developing prediction approaches that are based on estimations of dynamics and conformation directly from experimental NMR data. These estimations are less accurate but can encompass the whole range of protein behaviour, from intrinsically disorded via molten globule to fully folded. This is the basis of the DynaMine method published at the end of 2013. We have evidence that the predictions add a new, physical, dimension to the protein sequence that has many uses, such as improving sequence alignment or detecting early folding events – areas currently under investigation.
People
Taushif Khan, Gabriele Orlando, Daniele Raimondi, Wim Vranken